It has occurred to me that with all the new technology advances over the last five years that my programming skill set may be becoming obsolete. There has been much more emphasis on web page design than in, say, computational fluid dynamics; more cycles and (electronic) ink being spent on machine and deep learning than new parallelism support in Fortran 2015. Any new developments in the areas of computational science and technical computing have been dominated with languages like Python, R, Julia, and Scala. These are all good, of course, since they allow researchers and practitioners to be able to design and code powerful applications without needing the assistance of a computer scientist (like me). *sniff*
Let me give you an example from some code I’ve been working with. Part of the larger problem was to read in a list of edges (source vertex, destination vertex) describing a directed graph and find the contained undirected subgraph. That is, keep only those directed edges--and participating vertices—that had reciprocals. Another way to say that is to keep directed edge (a, b) if and only if the original graph also contained edge (b, a).
The first decision to make when designing an algorithm is to pick a data structure that supports the computation. One obvious idea is to use an adjacency matrix to represent the graph. If I see that the edge (a,b) is in the graph (G[a, b] == 1) then I can quickly check that the reciprocal edge is also in the graph (G[b, a] == 1) and place edges (a,b) and (b,a) into the resultant undirected graph’s adjacency matrix.
However, if there are millions of vertices in the graph and it is known that there are maybe 10-25 edges from each vertex, then I have a sparse graph and an adjacency matrix will be more than 99.999% zero entries. In such cases, it is a better use of memory resources to utilize some form of a compressed-sparse-row (CSR) storage scheme. One that I like uses one array of pointers where each element corresponds to a vertex. The pointer indexed by a vertex points into another array of vertex labels that are at the destination “end” of an edge with the vertex as the source. The list of “neighbors” for vertex n ends where the pointer for vertex (n+1)’s points into the destination array. I could probably write another thousand words about this, but here’s a picture to take the place of those words.
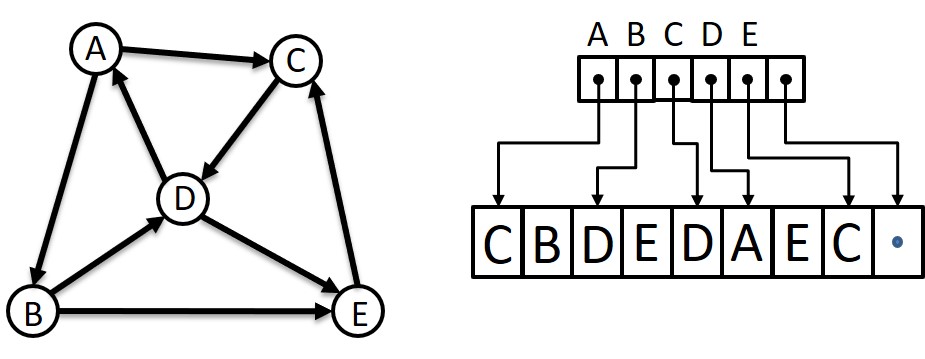
To find undirected edges an algorithm would take each vertex in turn, call it X, and for each destination vertex (or neighbor), call it Y, check the list of Y’s neighbors and include the edge (X, Y) if one of Y’s neighbors is X. The edge (Y, X) will be added to the undirected graph when processing Y as the source vertex. Thus, I would write a double-nested loop (for each vertex, for all neighbors of the vertex) with a test to match reciprocal edges and some way to add those edges to another graph structure holding the undirected graph being sought. This would be an O(n2) algorithm and I was content.
My contentment lasted until I saw the following source line from a Python code running under Spark to perform this graph operation:
edges = edges.intersection(edges.map(lambda x: (x[1], x[0])))After reading the graph’s list of edges into an RDD (resilient distributed dataset), named edges, a map operation is executed on every edge to reverse the order of the vertices, which creates a temporary RDD holding those reverse edges. The set of reversed edges is then intersected with the original set of edges, keeping only those items that are in both sets. Hence, only those edges that have both (X, Y) and (B, Y) in the original edge list will be stored in the resulting RDD since (Y, X) and (X, Y) would both be in the mapped set.
Elegant. Quite elegant.
In one line of Python code running in a Spark context, the same operation as my 6-8 lines of C, described above, is accomplished. And it can run that operation in parallel if there are processing resources assigned to the Spark environment. To make my version parallel I would need to add another line or two of OpenMP and also add code to synchronize access to the data structure holding the final set of reciprocal edges.
Looking at the above Python line, I began to feel almost like Captain Dunsel. Sure, there are issues of execution speed and memory usage and overall performance between my code and the Python that uses Spark libraries, but those are for another blog. I’m too dismayed to cite them, even if they might all be in my favor.
However, I have thought of two ideas where I might be able to avoid the scrap heap. The first is that there will need to be teachers of the methods of these new programming paradigms. Even if my abilities to code “near the metal” won’t be in much demand going forward, I know that the next generation of coders will need someone to point out and explain all the new, time-saving features of a system. Which brings me to my second idea: devising the overall algorithm to perform a complex computation in an elegant way. The above line of Python code to pull out the undirected subgraph from a larger directed graph isn’t inherently obvious, especially to someone not versed in Computer Science. (In some cases, like me, it wasn't immediately obvious to those that are steeped in CS.) So, since I do have a computational background and some decades of experience, I could be useful as an algorithmic consultant to put together the bits of arcane and semi-mystic operations into a tight, coherent code that will accomplish the larger task.
At least until they can teach a robot to do the same things I can.