Challenge
Genetics testing is reaching a stage where techniques for combatting diseases based on individual genetic information are becoming viable and discoveries in epigenetics are delivering breakthrough results in understanding the mechanisms of gene expression. Advanced pattern recognition and the latest breakthroughs in processing power have proven invaluable in evaluating massive databases of genetic information and pinpointing correlations. For example, in the project discussed in this story, epistasis detection is an important tool in bioinformatics studies, identifying associations in genotypes (collections of genes) and phenotypes (observable characteristics) on a genome-wide basis.
In basic terms, epistasis is the phenomenon in which one or more genes affect the expression of another gene, modifying it through masking, suppression, or inhibition. Single nucleotide polymorphisms (SNP) are genetic markers that interact with each other to affect complex traits or diseases, such as Alzheimer’s disease, diabetes, or various types of cancer. High-order epistasis can identify multiple SNP with correlations that are helpful in determining individual susceptibility to certain diseases, as well as personalizing treatment for these diseases.
Streamlining Detection
Two projects were combined to improve the prospects for efficient epistasis detection:
- Boosting Epistasis Detection on Intel® CPU and GPU Systems (hosted on Intel® DevMesh)
- Cache-aware Roofline Model with Intel® Advisor.1
Unlike typical performance-measurement models that use percent-of-peak estimates, the Cache-aware Roofline Model combines locality, bandwidth, and different parallelization paradigms into a single performance figure. Integrated with Intel Advisor, this model effectively analyzes whether an application is configured to achieve the best possible performance given the characteristics of the computer. This performance analysis can be extended to explore other algorithms with the goal of increasing their performance.
Rooflining High-Order Epistasis Detection
These research projects were conducted with the help of INESC-ID research institute.2 Headquartered in Lisbon, Portugal, INESC-ID is involved in many strong prospective projects with the funding obtained through the European Union and/or Portuguese Foundation for Science and Technology (FCT) (supported in this case by the FCT, Intel, and other parties). Projects typically proceed from a fundamental R&D level to latter stage training and strategic initiatives.
As part of a survey to discover associations between genotypes and phenotypes, the compute process is designed to take advantage of extensive parallelism to accelerate the pattern identification. Support for cross-platform application compatibility underlies the project—a fundamental benefit provided by oneAPI programming models and tools. Heterogeneous hardware components have become commonplace in today's computer systems, and there are clear advantages to simplifying programming tasks by using an open, cross-architecture approach. Code equipped to take advantage of available XPUs (CPUs, GPUs, FPGA, and all available accelerators in a system) can deliver optimal results on a diverse range of hardware platforms.3
Background
Aleksandar Ilic is an assistant professor with the Department of Electrical and Computer Engineering (ECE) of Instituto Superior Técnico (IST), University of Lisbon, Portugal. He received a PhD in ECE in 2014. Aleksandar brought his expertise and deep understanding of the Cache-aware Roofline Model to the project goals of accelerating epistasis detection using code that was compatible with a wide range of hardware platforms. Working with like-minded colleagues, Aleksandar and the team made notable advancements in bioinformatics research and performance measurement in complex systems composed of heterogeneous hardware.
Speaking about the adoption of the Cache-aware Roofline Model as the mechanism for the ensuring best performance to meet the project objectives, Aleksandar said, "The Cache-aware Roofline Model represents a novel take on the roofline modeling (when compared to the traditional approach). For the first time, it gave the possibility to visualize the bottlenecks of a complete memory hierarchy in a single plot. It also provided new interpretations, bottleneck detection and application optimization methodology."
In the following sample roofline chart:
- X-axis indicates the arithmetic intensity, measured in floating-point operations per byte (FLOP/byte).
- Y-axis indicates performance level in giga floating-point operations per second (GFLOPS).
- Dots show loops or functions in an application.
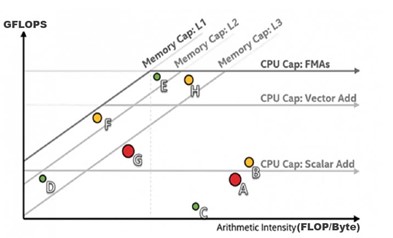
"Our recent contributions in epistasis detection," Aleksandar said, "represent one of the fastest solutions in the existing literature (surpassing some state-of-the-art approaches by several times)."
Solution
Methodology and Approach
A key development that became a cornerstone of the Epistasis Detection project took place in 2017 with the integration of Cache-aware Roofline Model as an official feature of Intel Advisor. This made it possible to build roofline plots and the bioinformatics application within a oneAPI framework across an extended expanse of Intel devices, covering the latest CPU and GPU microarchitectures.
"We rely on these insightful models in heterogeneous computing environments,” Aleksandar said, "to boost the execution of bioinformatics codes using oneAPI and DPC++ programming language. For this purpose, we focus on epistasis detection, which is a challenging and important bioinformatics application that may require processing of large volumes of data representing genetic markers such as SNP."
Intel Tools Used
Aleksandar and fellow researchers used Intel® Developer Cloud to construct the epistasis solution. "We used [Intel] Developer Cloud for development—a great free platform, which is certainly one of the most important driving vehicles behind the wider adoption of DPC++ (at least in the scientific community) and promotion of the latest Intel hardware."
As confirmed by recent study results, Aleksandar is convinced that the proposed Epistasis Detection project, which is based on the oneAPI framework and tools, delivers substantially better performance than prior approaches and provides the benefit of code portability that encompasses the CPU and GPU devices produced by the three leading companies in the market.
Linux* was used as the operating system throughout the coding work. Key Intel technologies used in the project include:
- Intel® oneAPI Base Toolkit
- Intel® oneAPI HPC Toolkit
- Intel® Parallel Studio
- Intel® VTune™ Profiler
- Intel® Developer Cloud
- Intel® Xeon processors
- Intel® GPUs
- Intel® FPGAs
Key Challenges Met
Bioinformatics, by nature, involves massive volumes of data, which can lead to vast combinations possibilities to evaluate. Optimizing application performance across diverse architectures requires being able to pinpoint execution bottlenecks, whether caused by hardware or software.
"For bioinformatics codes, a particular challenge that we experienced was providing code and performance portability across heterogeneous environments. For example, we were initially faced with a burden of deriving and optimizing the algorithms for different architectures, which required the use of vendor-specific programming models and tools (CUDA*, HIP, Intel intrinsics, and so on). Then SYCL (DPC++) appeared, which allowed us to easily port our codes to different architectures, while still not experiencing significant performance drawbacks."
To improve processing efficiency, the project team developed a dynamic scheduler to distribute the genotype combinations across available CPUs and GPUs. "For this purpose," Aleksandar said, "we derived a specific module for dynamic scheduling, where the workload distributions are decided at runtime. In other words, the distributions (consisting of the number of genotype combinations to be assigned to each device) are decided based on the performance estimations and measurements for each device, which are obtained from the previous execution rounds."
Research Accelerated by Enhanced Parallelism Techniques
Research into the high-order epistasis detection coincides with the rising understanding of the genetic basis of disease, but also presents challenges in terms of the complexity of the data involved and the difficult quest to pinpoint useful associations given the amount of data that must be ingested.
When asked if the project might offer value to other developers, Aleksandar responded, "I would like to believe our approach can help other researchers and developers to optimize their applications from many different areas. Some of the optimization techniques that we adopted for our bioinformatics codes can be helpful when parallelizing applications with similar execution patterns."
Conclusion
"Some of the best discoveries happen in unexpected moments (and after a lot of work)," Aleksandar said. "For example, our initial investigation of roofline modeling was not aimed at developing new models. It just happened during the research process in which we targeted the extension of the original model for energy efficiency."
Aleksandar offers this advice to other developers embarking on ambitious projects in innovative areas of research: "Take a deep breath—it’s a long run, and it can be frustrating. However, seeing the outcomes is very rewarding and it outweighs all those struggles."
Resources and Recommendations
Running a Roofline Analysis with Intel Advisor
Published by NASA*, Running a Roofline Analysis with Intel Advisor delivers insights into the performance-enhancing opportunities available using this type of performance analysis.
Cache-aware Roofline Model
A notable paper on a Cache-aware Roofline Model exposes its fundamentals and overall usability, while the follow-up papers document the implementation in Intel Advisor and its application-specific extensions.
More Information
Implement a Cache-aware Roofline Model in Intel Advisor
Application-driven Cache-aware Roofline Model
Boost Epistasis Detection on Intel CPU and GPU Systems
This Intel DevMesh project features research work on boosting epistasis detection on Intel CPU+GPU systems, to which Aleksandar Ilic was a key contributor, looks into the ways that Intel Advisor and Intel VTune Profiler can be used in combination with Data Parallel C++ (DPC++) to accelerate epistasis detection techniques on heterogeneous computer architectures.
Access the following contributions in the HiperBio GitHub* repository. The contributions resulted in several scientific publications, such as the following comprehensive references for understanding how to achieve parallel speedups on diverse microarchitectures:
- Unlocking Personalized Healthcare on Modern CPUs/GPUs: Three-way Gene Interaction Study
- Heterogeneous CPU and iGPU Processing for Efficient Epistasis Detection
Cross-architecture High-Order Exhaustive Epistasis Detection on CPU and GPU Devices
Hosted on Intel DevMesh, see how CPU and GPU nodes on Intel DevCloud proved valuable in using an application built with SYCL/DPC++ to perform complex searches or find correlations between genetic markers.
Intel Developer Cloud for oneAPI
Build and optimize oneAPI multiarchitecture applications using the latest optimized Intel® oneAPI and AI tools, and test your workloads across Intel® CPUs and GPUs. No hardware installations, software downloads, or configuration necessary. Free for 120 days with extensions possible.
1 This tool is available as a stand-alone component or through the Intel® oneAPI Base Toolkit, and provides a way to evaluate optimal use of available compute resources.
3 Highlights of the project can be found at Sparcity.edu.
The full paper is available as a PDF: Unlock Personalized Healthcare on Modern CPUs and GPUs
About the Author
Highlights of Aleksandar’s career include:
Senior researcher in the High-Performance Computing Architectures and Systems group (HPCAS) at INESC-ID.
Extensive experience with the Cache-aware Roofline Model, including over 20 roofline-related tutorials. He received the HiPEAC Tech Transfer Award for integrating this model into Intel Advisor and other industry software tools.
Substantial achievements in bioinformatics by developing high-performance and energy-efficient running of applications in this space. Several best paper nominations for this work have been extended at prestigious international conferences, including International Parallel and Distributed Processing Symposium (IPDPS) and Field-Programmable Custom Computing Machines (FCCM).
Contributions to international events, including talks, scientific venues, and seminars, as well as authoring and coauthoring over 60 papers for scientific journals and conferences worldwide.