Introduction
Image recognition with machine learning techniques has achieved significant growth due to advances in recent years in both algorithmic efficiency and hardware performance. Even with these advances, image pre-processing of raw images remains a critical step, especially in larger datasets. There are variety of methods to pre-process images, but the SIFT + Bag-of-Words (BoW) routine has gained popularity for its straight forward approach.
This article will discuss the application and comparison of SIFT + Bag-of-Words on two different data analytics machine learning libraries, scikit-learn* and Intel® Data Analytics Acceleration Library (Intel® DAAL). It will start with batch processing, then move on to utilize Intel DAAL distributed processing mode interfaced with the open-source mpi4py application. scikit-learn is a popular Python* module with a mature set of features and easy to use API. Its ease-of-use is well-known, but in some areas it lacks in speed and in the ability to distribute, which will serve as the basis for this article's comparison.1
Description of the SIFT and Bag-of-Words Routine
SIFT
SIFT (Scale-Invariant Feature Transform) algorithm is an emergent image processing technique used to identify important features in raw images and convert them to usable numerical format. SIFT detects interest points in an image, then transforms the points into both scale and rotationally invariant vectors containing 128 dimensions. The resulting vectors are called key descriptors. SIFT generates 0 to m descriptors for each image producing a matrix with m x 128 elements; where m represents the number of interest points identified by SIFT algorithm.
BoW Model
The bag-of-words model uses a binning of similar words, quantified by the number of occurrences within a document. A related approach called visual bag-of words is implemented in the present exercise to learn features based on the SIFT descriptors and the number of occurrences in image.
SIFT and BoW Model Implementation
As described in the SIFT section, SIFT features detected on each image are comprised of m x 128 key descriptors, where m is the number of interest points in an image. Images will have various m values based on the number of points the SIFT algorithm identifies as interesting. A full stacked list of key descriptors for all images (in) under study are gathered in a common feature space, where similar key descriptors are clustered with the k-means algorithm and subsequently assigned to a cluster or bin. Finally, a histogram of these bins are generated for each individual image resulting in a final matrix size of inx k.
Note on the Image Data Set Used
The dataset used for this work is an internal image set, contributed to by Intel employees. Photographs were taken around the Hillsboro, OR campus, as well as employee's homes. Examples of subjects are a person's shoes, bedroom pictures, and cubicle shots of keyboard, monitors, and chairs. The full set includes many examples of each type, for instance, 20 different sets of shoes. The image set was manually labeled into rough classes (shoes, chairs, monitors, etc..) and saved. There were 720 image files of 1920 x 1080 pixels, totaling ~0.6 GBs on disk. 4 million SIFT features vectors were extracted, each with 128 features. The authors are prohibited from showing the images to the public, as they are only for internal benchmarking and testing.
See Section A in Code Snippets demonstrating SIFT feature extraction using OpenCV library.
Tuning Number of Clusters k
Importance of Tuning k
As described above, the bag-of-words model collapses extracted SIFT features into a single feature vector by binning based on number of occurrences of a given type. Type in this case is assigned by using a clustering classifier. k-means is the most commonly used clustering algorithm for these purposes, and the quality of the clustering is governed by the number of k chosen during the fit. A downside of employing the k-means algorithm is that the user must supply the k (number of clusters) value before fitting. If k is too low, the resulting assignment and distance measurements will be weak. If k is too high, downstream work is more likely to bottleneck due to increased noise in the system. Often k is hard to assign without some trial and error. Basic search tuning is a common solution, where a user decides on a scoring function, and plots the score with various values of k. Figure 1 is a plot of k tuning, using the accuracy of a downstream Random Forest classifier as the scorer.
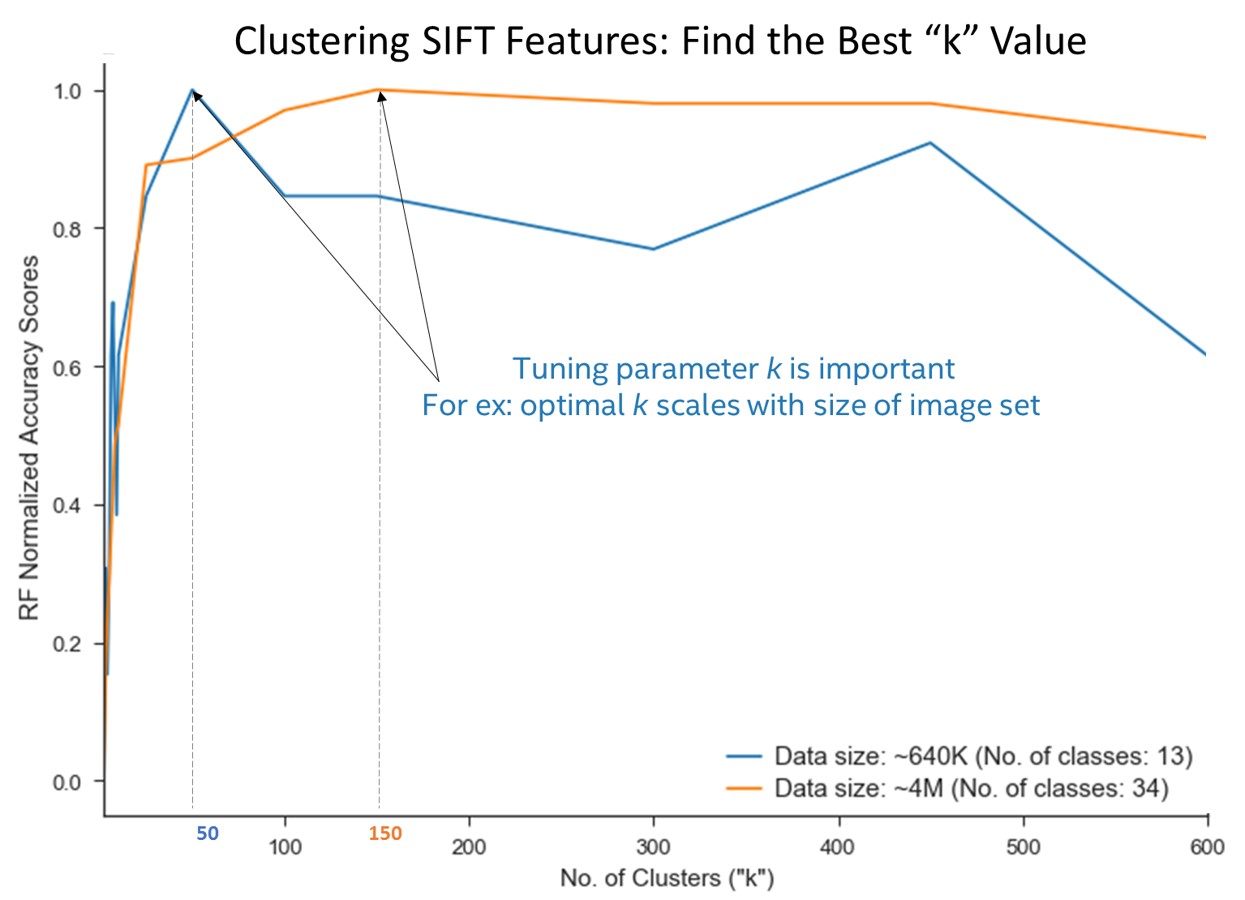
Figure 1. Random Forest (RF) score plotted against k value for fitting of k-means algorithm. Datasets consisted of 13 classes (640 K SIFT features vectors) and 34 classes (4 dillion SIFT features vectors), each with 128 features. The annotations at k values of 50 and 150.
See Hardware Spec 1 for hardware and configuration information
Figure 1 includes two different sized datasets, consisting of 13 classes (640 K SIFT features vectors) and 34 classes (4 million SIFT features vectors), each with 128 features. It is clear that each line plot presents an elbow where score is maximized, but cluster number is kept low. It is also clear that collection of these data points requires many k-means algorithm fits in succession. As such, speed and scalability of these fits is extremely important. Needless to say, if fitting on.
4M x 128 data set for 20 or 50 k took ~5-10 hours, tuning for k would be an unsustainable proposition. In addition, k-means is susceptible to local minima, so many users will fit multiple times for a given k.2
See Section B in Code Snippets demonstrating k-means cluster on SIFT features using Intel DAAL library. This code snippet converts m x 128 SIFT data set into a in feature vectors described by k features.
Feasibility of k Tuning: PyDAAL vs scikit-learn
A proper tuning of ML hyperparameters should include lead-room on either side of the optimal position to ensure avoidance of local minima. In other words, the more parameter positions visited during the tuning process, the more reliable the outcome for the user. To this end, tuning speed is the key to a sustainable approach to finding correct k values in the visual bag-of-words routine. Figure 2 below plots execution time vs k tuning results using PyDAAL and base-Python scikit-learn clustering implementations.
Figure 2. Tuning of k execution times (plotted on log scale) for PyDAAL and base-Python scikit-learn clustering implementations. Dataset consisted of 34 classes (4 million SIFT features vectors), with 128 features. The scikit-learn curve stops at k = 150 due to unsustainable execution time.
See Hardware Spec 1 for hardware and configuration information
Figure 2 shows result of tuning k with log execution time on the y-axis. The two series are results for PyDAAL (blue) and base-Python scikit-learn (orange) clustering implementations. The Dataset consisted of 34 classes (4 million SIFT features vectors), with 128 features. Figure 1 showed optimal k for this dataset at a value of 150. A proper parameter search for k should span positions above and below k = 150. The scikit-learn curve stops at 150 k due to an unsustainable execution time of ~26k seconds. Therefore a proper parameter search with scikit-learn would only be possible with days or weeks allotted for tuning of a single batch. For reference, the entire PyDAAL plot was collected in less time than the k = 150 data point on the scikit-learn series.
See Section C in Code Snippets demonstrating k-means cluster tuning using Random Forest Classifier. Accuracy metric is used to decide the best choice of k.
Distributed Tuning of k on Large Datasets
As the size of SIFT-extracted dataset rises, even the high performance PyDAAL implementation of k-means algorithm will become ill-fitted for proper fitting. At some point the one-shot, all-at-once fitting approach (aka batch) must be discarded and the distributed approach inserted. Scikit-learn is a mature library and a pioneer in usability and workflow enabling, but its lacks support for this sort of distributed fitting.
PyDAAL includes a host of classes, built especially for allowing the user to insert into current distribution infrastructure. PyDAAL provides methods for splitting the data into chunks, sending in serialized form, deserializing, partial fitting, and collective reduction for finalizing splits. The PyDAAL distributed methods work like primitives, and allow the user to use any communication strategy they desire. The binding module utilized in this exercise is Mpi4Py module which calls Intel MPI under the hood. The data collection below summarizes k tuning on a large dataset which consists of 16 million SIFT vectors with 128 features.
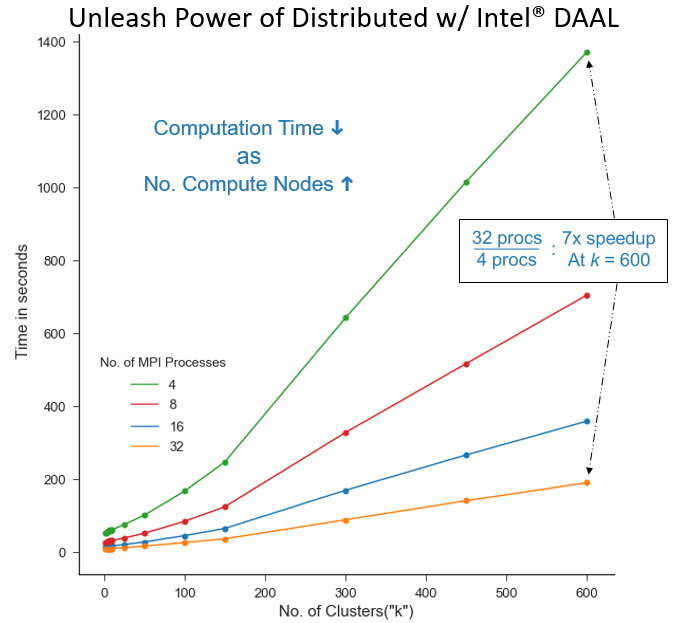
Figure 3. Demonstration of using PyDAAL in distributed mode to tune k for a large dataset. Intel MPI with Mpi4Py used for communication bindings. Each run had 2 MPI ranks per node and threaded MKL running on max threads available. Dataset consisted of 16 million SIFT feature vectors with 128 features.
See Hardware Spec 2 for hardware and configuration information
Figure 3 Demonstrates using PyDAAL in distributed mode to tune k for a large dataset (16 million SIFT features vectors with 128 features). The computation time of k-means fitting scales inversely with number of processors. For example the recorded data points for k = 600 at 4 and 32 MPI processes are 901s and 127s, respectively. That is a 7x speedup from distributing the computation among 8x processes. Considering the 8x speedup would be the theoretical maximum, PyDAAL's implementation is highly efficient. This allows Python programmers to pull maximum power out of their hardware clusters and lower the time it takes to tune hyperparameters, all without writing complicated control logic code.3
See Section D in Code Snippets demonstrating k-means clustering in distributed mode implementing Mpi4Py.
Conclusion and Call to Action
PyDAAL is a fast and powerful analytics library. An important advantage of PyDAAL over scikit-learn is in the computation time required during algorithm fitting. Often users need to tune hyperparameters by varying across a parameter space and scoring the output with some sort of quality metric. The increased speed of the each PyDAAL algorithm fit allows for more parameter space and increments in a user's grid search coverage. Furthermore, as datasets grow to exceed in-machine memory capacity, PyDAAL can be used to distribute calculations across multiple machines. PyDAAL's k-means distributed implementation is very efficient and adds speedup commensurate with the increased number of resources made available without the users writing complicated control logic. Get started with PyDAAL as a part of the free Intel® Distribution of Python today and feel free to harvest the code from the below code sections for your own work.
Appendix
Code Snippets
Prerequisites
Python: Intel® Distribution for Python*
Packages: Intel DAAL Update 1 or above, numpy, scikit-learn, opencv, mpi4py, customUtils (available in GitHub repo here), k-means helper class from here
Note that path variable from code section A must be the location to your <path> that has the directory structure <path>\category\image_files
How to Run the Snippets?
To experiment Batch processing, Run Code Sections A, B and C together as a single script
To experiment Distributed processing, Run Code Section A with additional codelines as shown below to save the SIFT array
import pickle as pk
pk.dump(all_img_sift_array,open('sift_dat.p', "wb"))
Save code section D as script.py and run the Linux* bash command as shown at the end of section D.
A. SIFT Feature Extraction from Images Using OpenCV*
import cv2
from glob import glob
from os.path import join,basename
from numpy import zeros, resize, histogram, vstack, zeros_like, append
def get_imgfiles(path):
all_files = []
for fname in glob(path + "/*"):
all_files.extend([join(path, basename(fname))])
return all_files
def extractSift(img_files):
img_Files_Sift_dict = {}
for file in img_files:
img = cv2.imread (file)
gray = cv2.cvtColor (img, cv2.COLOR_BGR2GRAY)
sift = cv2.xfeatures2d.SIFT_create()
kp, des = sift.detectAndCompute (gray, None)
img_Files_Sift_dict[file] = des
return img_Files_Sift_dict
def Siftdict2numpy(dict):
nkeys = len(dict)
array = zeros((nkeys * 1000, 128))
pivot = 0
for key in dict.keys():
value = dict[key]
try:
nelements = value.shape[0]
except AttributeError:
print("## Image file with 0 SIFT descriptors - {}".format(key))
value = np.zeros((1,128))
dict[key] = value
nelements = value.shape[0]
while pivot + nelements > array.shape[0]:
padding = zeros_like(array)
array = vstack((array, padding))
array[pivot:pivot + nelements] = value
pivot += nelements
array = resize(array, (pivot, 128))
return array
def SiftNumpy2Norm(all_img_sift_array):
from sklearn.preprocessing import MinMaxScaler, StandardScaler
scaler = StandardScaler()
return scaler.fit(all_img_sift_array).transform(all_img_sift_array)
all_img_sift_dict={}
all_files_labels_dict={}
path='' # directory structure is assumed of the form "\category\images'"
if path=='':
raise SystemExit("Error: Enter the location of . (directory structure is assumed of the form '\category\images'")
label = 0
for category in glob(path + "/*"):
img_files = get_imgfiles(category)
all_img_sift_dict.update(extractSift(img_files))
for i in img_files: all_files_labels_dict[i] = label
label+=1
all_img_sift_array = Siftdict2numpy(all_img_sift_dict)
all_img_sift_array= SiftNumpy2Norm(all_img_sift_array)
B. VisualBagOfWords with Intel® DAAL (batch): K-means + Binning
import time
from Kmeans import Kmeans #Kmeans daal class available in pydaal-getting-started GitHub repo
from numpy import histogram
from customUtils import getArrayFromNT#customUtils available in pydaal-getting-started GitHub repo
from daal.data_management import HomogenNumericTable_Float64
def computeVisualBoW(all_img_sift_array,all_img_sift_dict,all_files_labels_dict,nclusters):
def clustering(all_img_sift_array,nclusters):
all_img_sift_array_nT = HomogenNumericTable_Float64(all_img_sift_array)
kmeansPydaal = Kmeans(nclusters)
start = time.time()
KM = kmeansPydaal.compute(all_img_sift_array_nT)
print("Computation time taken to calculate centroids on {} via K-means: {:2f}s".format(nclusters,time.time()-start))
return KM
def createBins(nclusters, assignedClusters):
histogram_of_words, bin_edges = histogram(assignedClusters,
bins=range(nclusters + 1))
return histogram_of_words
KM = clustering(all_img_sift_array,nclusters)
centroids = KM.centroidResults
all_word_histgrams_dict={}
for imagefname in all_img_sift_dict:
assignedClusters_nT = KM.predict (centroids, HomogenNumericTable_Float64(all_img_sift_dict[imagefname]))
assignedClusters = getArrayFromNT(assignedClusters_nT)
word_histgram = createBins(nclusters,assignedClusters)
word_histgram = append(word_histgram,all_files_labels_dict[imagefname])
all_word_histgrams_dict[imagefname] = word_histgram
return all_word_histgrams_dict
nclusters = 2 # 'nclusters' is value of 'k' in Kmeans. This is just an example value
all_word_histgrams_dict = computeVisualBoW(all_img_sift_array,all_img_sift_dict,all_files_labels_dict,nclusters)
C. Tuning K-means with Grid Search and scikit-learn Random Forest as Downstream Scorer
import numpy as np
from sklearn.ensemble import RandomForestClassifier
from sklearn.model_selection import train_test_split
lclusters = [2,3,4,5,6,7,8,9,10,25,50,100,150,300,450,600]
for nclusters in lclusters:
all_word_histgrams_dict = computeVisualBoW(all_img_sift_array,all_img_sift_dict,all_files_labels_dict,nclusters)
inpData = np.array(list(all_word_histgrams_dict.values()))
X = inpData[:,0:inpData.shape[1]-1]
Y = inpData[:,-1]
X_train, X_test, y_train, y_test = train_test_split(X, Y, test_size=0.4, random_state=0)
RFC = RandomForestClassifier ( class_weight='balanced').fit(X_train,y_train)
print(RFC.score(X_test,y_test))
D. Distributed K-means with Intel® DAAL and Intel® MPI
#The following scripts runs only on Linux OS
##################SECTION D:K-MEANS CLUSTERING IN DISTRIBUTED MODE##############
import sys, os
sys.path.append(os.path.join(os.path.dirname(sys.executable),'share','pydaal_examples','examples','python','source'))
from mpi4py import MPI
from utils import printNumericTable
from daal.data_management import HomogenNumericTable_Float64
import daal.algorithms.kmeans as kmeans
import daal.algorithms.kmeans.init as init
from daal import step1Local, step2Master
from customUtils import serialize, deserialize#customUtils available in pydaal-getting-started GitHub repo
import pickle as pk
rank = MPI.COMM_WORLD.Get_rank ()
name = MPI.Get_processor_name ()
size = MPI.COMM_WORLD.Get_size ()
def computestep1LocalInit(partialnT, nclusters):
localInit = init.Distributed (step1Local,nclusters, partialnT.getNumberOfRows()*size, rank*(partialnT.getNumberOfRows()-1), method=init.randomDense)
localInit.input.set (init.data, partialnT)
partialInitResult = localInit.compute ()
serialpartialInitResult = serialize (partialInitResult)
return serialpartialInitResult
def computestep1Local(partialnT, nclusters,centroids,nIterations):
localAlgorithm = kmeans.Distributed(step1Local, nclusters,it == nIterations, method=kmeans.lloydDense, assignFlag=True)
localAlgorithm.input.set(kmeans.data, partialnT)
localAlgorithm.input.set(kmeans.inputCentroids, centroids)
partialResult = localAlgorithm.compute()
serialpartialResult = serialize (partialResult)
return serialpartialResult
def computeOnMasterNodeInit(serialPartialResult, size, nclusters):
global finalResult
masterInitAlgorithm = init.Distributed (step2Master, nclusters, method=init.randomDense)
for i in range (size):
partialInitResult = deserialize (serialPartialResult[i])
masterInitAlgorithm.input.add (init.partialResults, partialInitResult)
masterInitAlgorithm.compute ()
finalInitResult = masterInitAlgorithm.finalizeCompute ()
return finalInitResult
def computeOnMasterNode(serialPartialResult, size, nclusters):
masterAlgorithm = kmeans.Distributed(step2Master, nclusters, method=kmeans.lloydDense,assignFlag=True)
for i in range (size):
partialResult = deserialize (serialPartialResult[i])
masterAlgorithm.input.add(kmeans.partialResults, partialResult)
masterAlgorithm.compute()
finalResult = masterAlgorithm.finalizeCompute()
return finalResult
all_img_sift_array=pk.load(open('sift_dat.p', "rb"))
serialPartialInitResults, serialPartialResults = [0] * size, [0] * size
sizeOfChunk = int (all_img_sift_array.shape[0]/ size)
rowStart = rank*sizeOfChunk
rowEnd = ((rank+1)*sizeOfChunk)-1
partialArray = all_img_sift_array[rowStart:rowEnd, :]
partialnT = HomogenNumericTable_Float64 (partialArray)#Partial inout data(numpy array) at each rank
#lclusters = [2,3,4,5,6,7,8,9,10,25,50,100,150,300,450,600]
lclusters=[10]
import time
for nclusters in lclusters:
#***************Initial centroid computation begins**************
if rank == 0:
start = time.time()
'''
**master routine**
computes partial init centroids on local
receives partial init centoids from all slave nodes(tag = 3301*)
computes final init centoids
sends final init centroids to all slave nodes(tag = 6602*)
'''
#Initializing lists to receive slave results
serialPartialInitResults,serialPartialResults= [0] * size, [0] * size
print("Master computing partial initial centroids")
serialPartialInitResults[rank] = computestep1LocalInit (partialnT,nclusters)
if size > 1:
print("Master receiving partial initial centroids from (tag = 3301*) from all slaves")
for i in range (1, size):
size, name, serialPartialInitResults[i] = MPI.COMM_WORLD.recv (source=i, tag=3301+nclusters)
result = computeOnMasterNodeInit (serialPartialInitResults, size,nclusters)
print("Master computing final initial centroids")
centroids = result.get(init.centroids)
print("Final initial Centroid {}".format(centroids.getNumberOfRows()))
serialCentroids = serialize(centroids)
if size > 1:
print("Master sending final initial centroids to all slave nodes(tag = 6602*)")
for i in range(1,size):
MPI.COMM_WORLD.send (serialCentroids, dest=i, tag=6602+nclusters)
else:
'''
**slave routine**
computes partial init centroids on local
sends partial init centroids to master(tag = 3301*)
'''
print("Slave computing partial initial centroids")
serialPartialResult = computestep1LocalInit (partialnT,nclusters)
MPI.COMM_WORLD.send ((size, name, serialPartialResult), dest=0, tag=3301+nclusters)
print("Slave sending partial initial centroids to master (tag = 3301*)")
#***************Initial centroid computation ends**************
#***************Final entroid computation begins**************
nIterations = 300##maximum no. of iterations
for it in range(nIterations):
'''
**master routine**
master computes partial centroids on local
receives partial centroids from all slave nodes(tag = 9903*)
sends final centroids to all slave nodes(tag = 13204*)
'''
if rank == 0:
if size > 1:
print("Master receiving partial centroids from all slaves (tag = 9903*)")
for i in range (1, size):
size, name, serialPartialResults[i] = MPI.COMM_WORLD.recv (source=i, tag=9903+nclusters+it)
print("Master computing partial final centroids")
serialPartialResults[rank] = computestep1Local(partialnT, nclusters,centroids,nIterations )
print("Master computing final centroids")
finalResult= computeOnMasterNode(serialPartialResults, size, nclusters)
centroids = finalResult.get(kmeans.centroids)
if nIterations > 1 and it < (nIterations-1) and size > 1 :
serialCentroids = serialize(centroids)
print("Master sending final centroids to all the ranks to compute partial centroids (tag = 13204*) for remaining iterations")
for i in range(1,size):
MPI.COMM_WORLD.send (serialCentroids , dest=i, tag=13204+nclusters+it+1)
else:
'''
**slave routine**
receives init centroids from master(tag =6602*) for first iteration
receives final centroids from master(tag = 13204*) if iterations>1 for further computation
computes partial centroids on local
sends partial centroids to master(tag = 9903*)
'''
if it==0:
print("Slave recieving final initial centroids from master (tag = 6602*)")
serialCentroids= MPI.COMM_WORLD.recv (source=0, tag=6602+nclusters)
else:
print("Slave recieving centroids from master (tag = 13204*)")
serialCentroids = MPI.COMM_WORLD.recv (source=0, tag=13204+nclusters+it)
centroids = deserialize(serialCentroids)
print("Slave computing partial centroids")
serialPartialResults= computestep1Local(partialnT, nclusters,centroids,nIterations )
print("Slave sending partial centroids to master(tag = 9903*)")
MPI.COMM_WORLD.send ((size, name, serialPartialResults),dest=0, tag=9903+nclusters+it)
if rank ==0:
printNumericTable(centroids,"Final centroids")
print("Computation time taken to calculate {} centroids via Kmeans: {:3f}s".format(nclusters,time.time()-start))
Linux bash command to Run
No. of MPI processes = No. of data partitions
mpirun -n 4 python script.py
Hardware Specs and Other Configuration Information
- Configuration Info - Versions: Intel® Data Analytics Acceleration Library 2018, scikit-Learn 0.19.0; Hardware: Intel® Xeon® Processor E5-2698 v3, 16 core CPUs (40MB LLC, 2.3GHz) 32 total threads, 32GB DDR4 RAM; Operating System: Ubuntu* 16.04.3 LTS.
Software and workloads used in performance tests may have been optimized for performance only on Intel microprocessors. Performance tests, such as SYSmark* and MobileMark*, are measured using specific computer systems, components, software, operations and functions. Any change to any of those factors may cause the results to vary. You should consult other information and performance tests to assist you in fully evaluating your contemplated purchases, including the performance of that product when combined with other products.
- Configuration Info - Versions: Intel® Data Analytics Acceleration Library 2018, scikit-Learn 0.19.0; Hardware: Intel® Xeon® Gold 6148 v3, 20 core CPUs (27.5MB L3, 2.4GHz) 40 total threads per node, 192GB 32GB DDR4 RAM; Operating System: Oracle* Linux Server 7.4 Kernel 3.10.0.
Software and workloads used in performance tests may have been optimized for performance only on Intel microprocessors. Performance tests, such as SYSmark and MobileMark, are measured using specific computer systems, components, software, operations and functions. Any change to any of those factors may cause the results to vary. You should consult other information and performance tests to assist you in fully evaluating your contemplated purchases, including the performance of that product when combined with other products.
References
- Intel DAAL has a fully functional Python API module called PyDAAL, which we will use for all of the code in this article. See article Gentle Introduction to PyDAAL: Vol 1 of 3 Data Structures. A companion article is available demonstrating use of new high-level Python API for Intel DAAL on the same workflow.
- The cost function for k-means fitting scales linearly with k.
- Intel MPI with Mpi4Py were used to run on 2 MPI ranks per node, and threaded MKL running on max threads available.