Executive Summary
This white paper provides an overview of fast Fourier transform (FFT), convolution, their application in medical image reconstruction, and gives example code showcasing the use. Here, medical imaging has been confined to computed tomography (CT), magnetic resonance imaging (MRI), and positron emission tomography (PET). However, similar concepts could apply to other modalities and respective image reconstruction procedures. This paper is for a beginner technical audience and provides a glimpse of FFT and convolution in the medical imaging domain.
Convolution
Convolution is a mathematical operation that is fundamental to various image-processing activities. In his talk, V. Hlavac1 describes convolution as an integral mixing of two functions where one function is overlaid and shifted over another. A continuous case convolution of two functions f and h can be mathematically represented the time domain as follows.
![]()
Where f is the input signal, h can be referred as a kernel, t is time, tau is the shift in time, and the asterisk symbol is usually used to represent convolution. In image processing, convolution provides a way of multiplying together two arrays of numbers of the same dimensions4 (for example 1D or 2D); however, they can be of different sizes (for example 3x4 convolved with 20x30). Convolution for 1D and 2D signals is described in detail in later sections in this white paper. Note that in the white paper integration is used for all continuous use cases and for discrete use cases, summation is used.
Convolution versus Cross-Correlation
Convolution and cross-correlation are similar operations with slight differences. As explained by R. Draelos in her article17, convolution means sliding a flipped kernel h across a signal f, while cross-correlation means sliding a kernel h across a signal f. Here flipping of a kernel means flipping values across all axes. For example, for a 2D signal, it would mean flipping both x and y axes (i.e., flipping top and bottom and flipping left and right).17
For a signal f and kernel h, in the continuous use case, convolution is represented with an asterisk and denoted as:
![]()
Cross-correlation is represented with a star as shown below. The bar on h represents flipping the kernel.
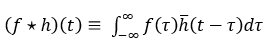
Image-Processing Convolution versus Deep Neural Network Convolution
It is important to understand the difference between convolution and cross-correlation because the word convolution in image processing can be confused with convolution happening between layers in a deep neural network in artificial intelligence (AI) applications.
In image processing, convolution occurs between an input array, which is normally an image, and a second much smaller array, sometimes called a kernel. Convolution is used for blurring images, sharpening, embossing, edge detection, and more5.
In deep learning, convolutional neural networks (CNNs) are a class of deep learning networks. CNNs consist of an input layer, an output layer, and, in between the two, several hidden layers. These hidden layers of a CNN usually have a series of convolutional layers that convolve with multiplication or other dot product6. These layers are popularly called convolutions; however, technically they are sliding dot products or cross-correlation. As R. Draelos mentions in her article17, most CNNs implement cross-correlation, but even if convolution was used instead, the same weight values would be learned by the network, just in a flipped orientation. In this paper, we refer to convolution as in digital image processing, so this convolution will not be confused with the recently more popular topic of CNNs in AI.
Fourier Transform
In his talk, V. Hlavac1 describes Fourier transform as one of the most commonly used signal processing techniques allowing a one-to-one transform of signal from the time-domain 

 f(t)
f(t) to the frequency domain F(ω) or vice versa.
to the frequency domain F(ω) or vice versa.
Mathematically, a Fourier transform can be represented as:
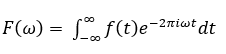
The inverse Fourier transform can be represented as:
![]()
A Fourier transform (FT) is usually applied to periodic signals. For non-periodic signals, a windowed FT or other methods are applied. It is worth noting that a weighted sum (integral) of many different complex exponentials can be used to express any 1D (dimension) function. In addition, as V. Hlavac1 mentions, since digital images are limited and have a finite number of discontinuities, Fourier transform always exists for digital images and hence applicability of FT in digital medical imaging too.
Discrete Fourier Transform
Discrete Fourier transform (DFT)2 is a numerical variant of the Fourier transform (FT), where discrete-time data is converted into discrete-frequency data sets. Since the resultant output is discrete, it is common for digital systems to use DFT when calculations are needed in the frequency domain.
If a continuous time signal f(t) is sampled every "T" seconds then one can obtain a discrete time signal f'(n). With the N samples taken every T seconds, the DFT can be computed over the compiled discrete time signal from the samples3.
If F(k) is the resultant discrete frequency spectrum of signal f'(n), then the DFT can be represented as:
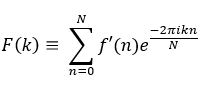
Here, f is used to denote a continuous time or spatial domain signal, f' denotes a discrete time or spatial domain signal and F denotes a signal in frequency domain.
Discrete-Space Fourier Transform
The discrete-space Fourier transform (DSFT)4 is used to transform two-dimensional (2D) signals that are discrete in the spatial domain to a continuous function in the frequency domain.
If f'(m,n) is a discrete signal in the spatial domain, then its DSFT to convert to the frequency domain is given as:
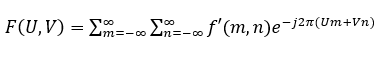
The inverse discrete-space Fourier transform, to go from the frequency domain to the spatial domain, is given as:
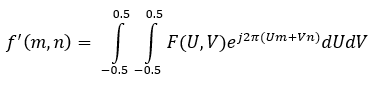
The forward DSFT is a summation because of the discrete nature of the spatial signal, but the inverse DSFT is an integral because the signal becomes continuous in the frequency domain. The fast Fourier transform (FFT), which is detailed in next section, is a fast algorithm to calculate the DFT, but the DSFT is useful in convolution and image processing as well.
As the Convolution Theorem18 states, convolution between two functions in the spatial domain corresponds to point-wise multiplication of the two functions in the frequency domain. An advantage of the DSFT is its convolutional linearity. Performing multiplication in the frequency domain after using the DSFT to transform the 2D signal or image will produce a linear convolution.4
Linear convolution of 2D signals or images involves dragging the filter over the image and computing the sum of the products of the image and filter only where they overlap. Circular convolution, however, involves dragging the filter over the image and the filter is periodically repeated all around itself until it overlaps the entire image. The sum of the products of the image and overlapping repeated filters is the computed convolution.4
Using the discrete-space Fourier transform for linear convolution is straightforward because there are no adjustments necessary to convolve the signal or image. However, it is still possible to perform a linear convolution on an image using the discrete Fourier transform.
Given a 2D image of size MxN and a filter or kernel of size PxQ, then linear convolution using the DFT and pairwise multiplication can be done by padding both the image and the kernel with zeros. When the image and kernel are both of size (M + P - 1) x (N + Q - 1), with zeros bordering the image and kernel on the right and bottom sides, then convolution using the DFT will be linear. This concept is demonstrated in the code examples in a later section.
Fast Fourier Transform (FFT)
When an image [input signal] and a kernel [impulse signal] are present in the time domain, convolution in the time domain can be used to obtain the resultant output. However, as described in the previous section, the mathematical implementation of convolution is complex and with larger datasets it gets more complex and time intensive. Another way to perform a similar operation on the signals and get the same output is to apply Convolution Theorem18 concepts and replace the time domain convolution with two DFTs to convert to the frequency domain, point-wise multiplication in the frequency domain, and then inverse DFT to return to the time domain. As mentioned by S. Smith in chapter 9 of his digital signal processing book7, this method reduces the large number of multiplications and additions involved in the time domain.
Using DFT and performing calculations in the frequency domain helps; however, for a discrete signal with N samples, the computational complexity, which directly impacts computational speed, for DFT is of the order of N2 as defined by V.Hlavac1. This computation speed issue can be resolved by using fast Fourier transform (FFT). FFT is a clever and fast way of implementing DFT. By using FFT for the same N sample discrete signal, computational complexity is of the order of Nlog2N . Hence, using FFT can be hundreds of times faster than conventional convolution7. Therefore, FFT is used for processing in the medical imaging domain too. Details of FFT are not provided in this white paper and readers are recommended to check chapter 12 of the digital signal processing book by S. Smith8 to learn more about FFT.
The Importance of FFT and Convolution in Medical Imaging
The above sections give an overview of convolution and Fourier transform as well as the mathematical and computational complexity involved with the two concepts. In this section, we will see how those convolution and Fourier transform concepts apply to the medical imaging domain and how critical these functions are in the field.
There are several modalities in medical imaging and the physics behind how an image is acquired and processed varies across each of the modalities. To keep the discussion focused, we will be mentioning the usage of FFT and convolution only in three medical imaging modalities, namely Computed Tomography (CT), Magnetic Resonance Imaging (MRI), and Positron Emission Tomography (PET).
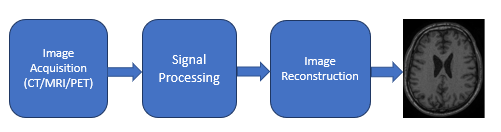
Figure 1: High-level block diagram for a generic (modality independent) image reconstruction pipeline, without data storage and other intermediate steps. Final image is of a brain MR, dataset 19
The image reconstruction block in Figure 1, is where FFT and convolution are heavily used in these modalities. The algorithm for image reconstruction could be different and specific to the device manufacturers but each of them usually uses FFT and convolution. In his report9, N. Abbasi details how FFT is used in CT. Hansen and Kellman in their publication10 explain how FFT is used in MRI image reconstruction. And the authors of image reconstruction in PET scanners11 explain how Fourier transform and convolution are used in PET imaging. For an overview of various image reconstruction pipeline algorithms in the three modalities and deployment of those by some device manufacturers, the authors recommend the presentation by M. Hansen on "Nuts & bolts of advanced imaging : the image reconstruction pipeline"12 as well as chapter 25 of S. Smith's book13 to better understand the role of FFT and convolution in various imaging techniques. Below we describe the relevance of FFT and convolution in CT imaging.
According to iData Research15, over 75 million CT scans are performed in the US each year. Each of these scans are running some FFT and convolution-based algorithm for image reconstruction. In order to give readers an idea of the extent of FFTs and convolutions occurring in medical imaging, let's take a deeper look on CT image reconstruction process.
As shown in Figure 2 (A), during CT data acquisition X-rays from a source pass through a body to a detector to obtain one view. The remaining views are obtained by rotating the source and detector inside the CT gantry in specific radial increments, until a complete 360 degree rotation is completed to obtain one image slice comprised of those views. Each sample in a view is the summation of all values along the ray that points to that sample. View 1 in Figure 2 (B) will be the sum of all values in each row at that specific fixed x-axis coordinate, while view 3 will be sum of all values in the columns for that specific y-axis coordinate. According to the Society of Imaging Informatics in Medicine14, on average a typical CT image is 512x512x12 bits. Per S. Smith13, in order to construct a 512x512 image slice a system might take about 700 views with 600 samples in each view. There are several algorithms to reconstruct an image slice given the views and samples in each view. One of the popular algorithms is called Fourier reconstruction13. In Fourier reconstruction, as S. Smith mentions13, first a 1D FFT is taken of each view, therefore requiring approximately 700 1D FFTs for a 512x512 image slice21. Secondly, these frequency domain view spectra are then used to calculate the 2D frequency spectrum of the image using convolution and Fourier slice theorem, requiring ~700 convolutions. Thirdly, an inverse FFT is taken of the spectrum to obtain the reconstructed image in the time domain using filtered back-projection technique as shown in Figure 2 (C), which requires an additional ~700 FFTs. By rough approximation this requires a total of about 1400 FFTs and 700 convolutions (excluding the convolution in filtered back-projection to be conservative) per 512x512 image slice21.
A CT machine can produce 64, 128, 256 et cetera slices, and the number of slices needed for a study will completely depend on the physician21. Using a 64-slice CT machine, producing a 512x512 image or slice, with each slice on average having 700 views, then based on the explanation above, the total number of FFTs would be approximately 89000 (64*1400) and the convolutions count would be approximately, 44800 (64*700). A larger study like CT angiography and cardiac CT14 will have a much higher number of views even for the same CT slices machine, hence a higher number of FFTs and convolution would be needed in those views.
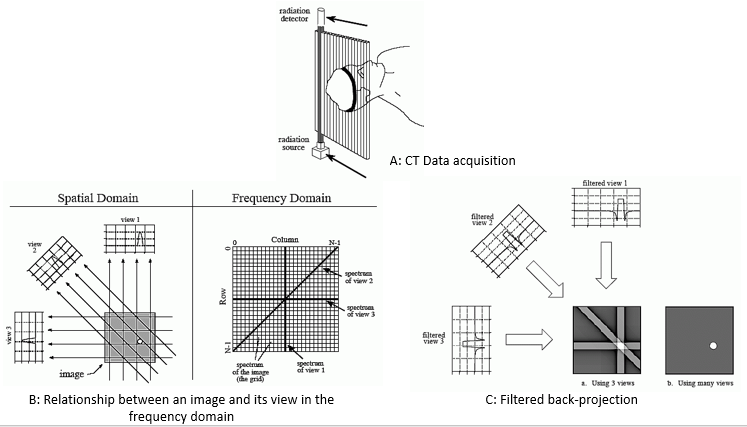
Figure 2: Major steps in CT image reconstruction using Fourier reconstruction method. Images taken from chapter 25 of S. Smith's book13
2D FFT and Convolution Code Example
This section provides some example 2D FFT and convolution C++ code snippets that take in a 2D gray scale image and convolve it with a 2D filter. The code shows two ways of performing the whole process. Method 1, which is referred to as brute force in the code, computes convolution in the spatial domain. Method 2 uses optimized built-in FFT functions from the Intel® Math Kernel Library (Intel® MKL)20 to compute the convolution of the image and the filter in the frequency domain. Intel® Math Kernel Library (Intel MKL) provides optimized FFT operations to take full advantage of hardware features in Intel® products including optimizing the implementation to maximize use of efficient hardware instructions.
For small data sizes, the brute force method can be optimal, however as datasets get larger, the FFT method is faster25. The output from either method is the same and users can choose to deploy the method they like.
Intel MKL also provides optimized functions to compute convolution. These functions automatically determine whether the brute force or the FFT method of computation would be faster and react accordingly.
The purpose of this demo is to exhibit the process of calculating the convolution of two signals.
Method 1: Brute Force Convolution
The "brute force" algorithm computes the convolution using the definition of convolution and not the fast Fourier transform. It drags the filter matrix over the image. At every pixel on the image, the values of the filter are multiplied by the corresponding values of the image that the filter is over. The products are summed, and this sum becomes the value of the pixel on the output image.
Here is a snippet of code from our demo showing the brute force convolution algorithm:
for(int i=0; i < rows; ++i) // rows
{
for(int j=0; j < cols; ++j) // columns
{
int out_index = i*s2 + j;
for(int m=0; m < kRows; ++m) // kernel rows
{
int mm = kRows - 1 - m; // row index of flipped kernel
for(int n=0; n < kCols; ++n) // kernel columns
{
int nn = kCols - 1 - n; // column index of flipped kernel
// index of input signal, used for checking boundary
int ii = i + (kCenterY - mm);
int jj = j + (kCenterX - nn);
int img_index = ii*s4 + jj*s1;
int kern_index = mm*s4 + nn*s1;
// ignore input samples which are out of bound
if( ii >= 0 && ii < rows && jj >= 0 && jj < cols )
out[out_index] += input_image[img_index] * kernel[kern_index];
}
}
}
}
s1, s2, and s4 represent the strides26 of the provided image and kernel and the output image. The variables kCenterY and kCenterX represent the indices of the center of the kernel in the Y direction and in the X direction, respectively. The code above assumes strides for the image and the kernel are the same.
Method 2: Convolution Using the Fast Fourier Transform (FFT)
To do convolution using the FFT, we use the Intel® Math Kernel Library (Intel MKL)20 to compute the FFT.
//create descriptor, configure settings, then compute the forward FFT
status = DftiCreateDescriptor(&my_desc1_handle, precision, DFTI_REAL, 2, input_sizes);
status = DftiSetValue(my_desc1_handle, DFTI_PLACEMENT, DFTI_NOT_INPLACE);
status = DftiSetValue(my_desc1_handle, DFTI_CONJUGATE_EVEN_STORAGE, DFTI_COMPLEX_COMPLEX);
status = DftiSetValue(my_desc1_handle, DFTI_INPUT_STRIDES, input_strides);
status = DftiSetValue(my_desc1_handle, DFTI_OUTPUT_STRIDES, output_strides);
status = DftiCommitDescriptor(my_desc1_handle);
status = DftiComputeFOrward(my_desc1_handle, pr2cFFT, pFFTResult);
//the computed forward FFT of our input image is now in the buffer pFFTResult
By the end of this snippet, the array pFFTResult holds the values of the image after going through the Fourier Transform. This function is then run again to compute the Fourier Transform of the filter, where output is stored in the array pImpulseFFTResult. Both the image and filter are transformed into the frequency domain after going through the FFT. In the frequency domain their values are complex numbers with real and imaginary parts. Point-wise multiplication on the real and imaginary values is accomplished as follows:
//do the elemenwise multiplication of the input image and the filter image
for (int i =0; i < diml; i++){
for (int j = 0; j < dim2; j++) {
int index = i*s2 + j;
temp = (pFFTResult[index].real * pImpulseFFTResult[index]2.real) - (pFFTResult[index].imag * pImpulseFFTResult[index].imag);
pFFTResult[index].imag = (pFFTResult[index].real * pImpulseFFTResult[index].imag) + (pFFTResult[index].imag * pImpulseFFTResult[index].real);
pFFTResult[index].real = temp;
}
}
To get the image back into the spatial domain, compute the inverse FFT, once again using the Intel MKL FFT functions.
The complete C++ code demonstrating the application of FFT and convolution to blur or sharpen a 2D gray scale MR brain scan image can be found on Gitlab. The code was run on the Intel® DevCloud for oneAPI23 and is available for the user to run through command line or through Jupyter* Notebook. Intel DevCloud for oneAPI provides access to the latest generation of the Intel® Xeon® Scalable processors, pre-installed and configured Intel® optimized libraries, compilers, and toolkits with a Linux* operating system terminal interface. Readers can register, sign in, and access the Intel DevCloud for oneAPI any time to run and test the code using Intel hardware and optimized software.
Conclusions
The authors would like to appreciate contributions made by Adina Golden to this white paper. The authors would like to thank their readers for investigating the white paper and the example code. FFT and convolution are just two of the many compute-intensive calculations occurring in a typical medical image reconstruction pipeline. The white paper provides insight into the computation behind a medical image reconstruction pipeline and is in no way comprehensive. There are multiple extensions of this project that the authors plan to work on and publish in due time.
References
- Fourier transform, in 1D and in 2D
- Digital Signal Processing or Discrete Fourier Transform: Wikibooks
- Machine Vision Study Guide: Frequency Domain Filtering
- 2.3 Basic Tools for Image Fourier Analysis
- Image Processing Learning Resources: HIPR2
- 3x3 Convolution filters
- Convolutional Neural networks: wikipedia
- Chapter 9: The Scientist & Engineer's Guide to Digital Signal Processing
- Chapter 12: The Scientist & Engineer's Guide to Digital Signal Processing
- The application of Fourier analysis in solving the Computed Tomography(CT) inverse problem
- Image reconstruction: an overview for clinicians
- Image reconstruction for PET scanners
- Nuts & Bolts of advanced imaging
- Chapter 53: Special imaging techniques
- Society of Imaging Informatics in Medicine
- iData research
- Machine Learning: Convolution vs Cross-correlation
- Convolution Theorem Wolfram Alpha
- Calgary-Campinas Public MR Brain Dataset
- Intel® Math Kernel Library
- Zwanger-Pesiri Radiology: What is a slice in a CT scan
- Periodicity of Inverse DSFT
- Intel®oneAPI DevCloud
- Intel® DevMesh
- FFT convolution vs direct convolution
- Image Stride